Transform a DAG object into a tidy_dagitty object
as_tidy_dagitty.DAG.RdThis function extends the as_tidy_dagitty function from the ggdag package to allow the input of a DAG object. The result is a tidy_dagitty object that includes only the structure of the DAG, without any specifications. May be useful to plot DAGs using the ggdag package. Note that completely unconnected nodes (no arrows going in or out) are ignored by this function.
Usage
# S3 method for class 'DAG'
as_tidy_dagitty(x, include_root_nodes=TRUE,
include_td_nodes=TRUE, include_networks=FALSE,
seed=NULL, layout="nicely", ...)Arguments
- x
A
DAGobject created using theempty_dagfunction with nodes added to it using the+syntax. See?empty_dagor?nodefor more details. Supports DAGs with time-dependent nodes added using thenode_tdfunction. However, including such DAGs may result in cyclic causal structures, because time is not represented in the output matrix.- include_root_nodes
Whether to include root nodes in the output matrix. Should usually be kept at
TRUE(default).- include_td_nodes
Whether to include time-dependent nodes added to the
dagusing thenode_tdfunction or not. When including these types of nodes, it is possible for the adjacency matrix to contain cycles, e.g. that it is not a classic DAG anymore, due to the matrix not representing the passage of time.- include_networks
Whether to include time-fixed networks added to the
dagusing thenetworkfunction or not. Usually it does not make sense to include those, because they are not classical nodes.- seed
A numeric seed for reproducible layout generation.
- layout
A layout available in
ggraph. Seecreate_layoutfor details.- ...
Optional arguments passed to
ggraph::create_layout().
Examples
library(simDAG)
# some example DAG
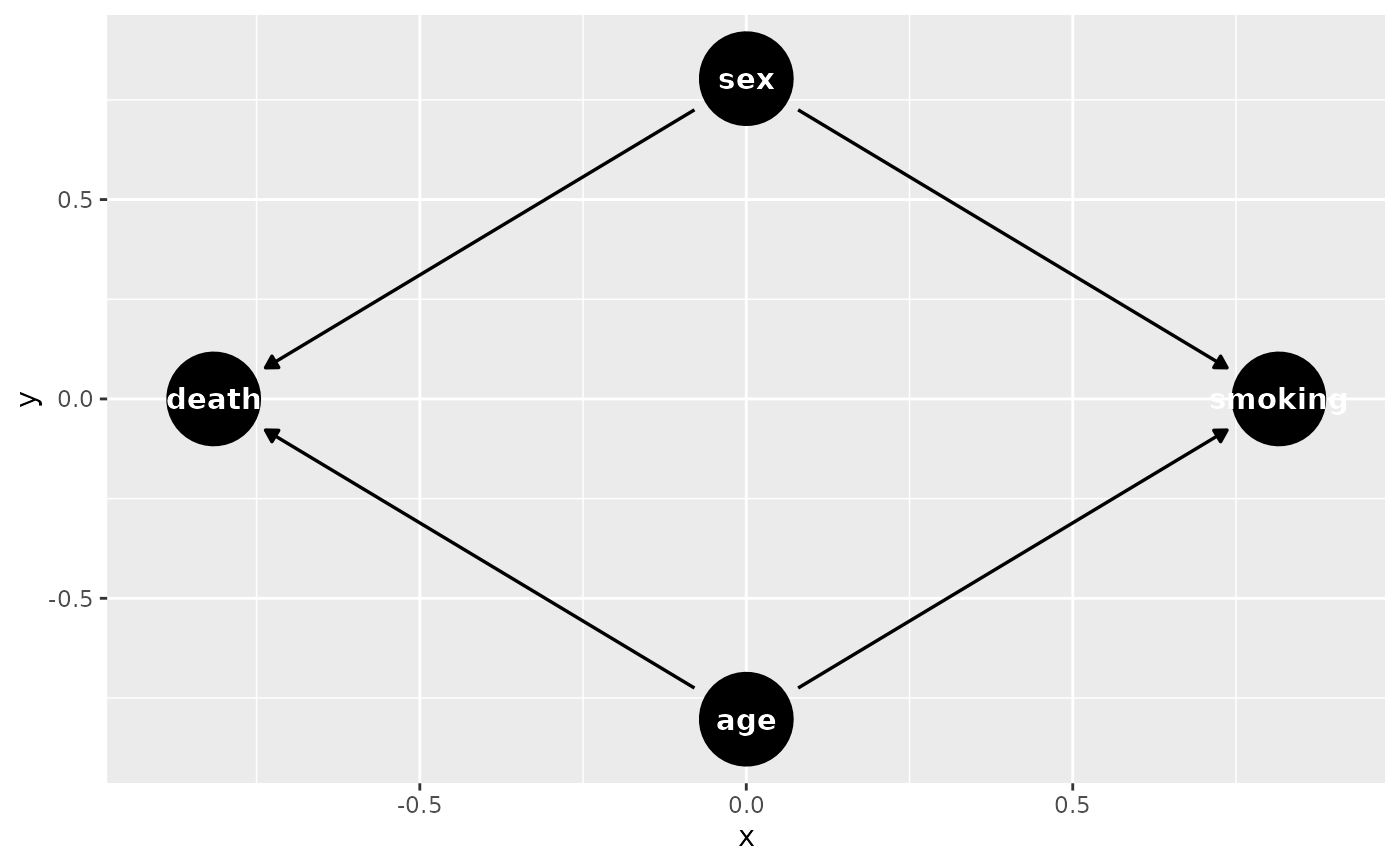
dag <- empty_dag() +
node("death", type="binomial", parents=c("age", "sex"), betas=c(1, 2),
intercept=-10) +
node("age", type="rnorm", mean=10, sd=2) +
node("sex", parents="", type="rbernoulli", p=0.5) +
node("smoking", parents=c("sex", "age"), type="binomial",
betas=c(0.6, 0.2), intercept=-2)
if (requireNamespace("ggdag")) {
library(ggdag)
g <- ggdag::as_tidy_dagitty(dag)
ggdag(g)
}
#>
#> Attaching package: ‘ggdag’
#> The following object is masked from ‘package:stats’:
#>
#> filter